Pubmedwhack: Immuno-DTO
Today’s Pubmedwhack comes from the world of unstable metals and electron microscopy. For the definition of “Pubmedwhack”, see this earlier post.
* * *
Following the invention of immunofluorescence, scientists developed other methods for using labeled antibodies to identify certain proteins or substances under a microscope.
There’s immunohistochemistry (IHC), in which the antibodies are labeled not with a fluorescent marker, but with an enzyme which produces a visible reaction in the presence of a substrate. This is less precise, but lets you see your protein of choice under visible light while also looking at the structure of the tissue. The slides labeled this way last longer instead of being quenched by the microscope’s light source.
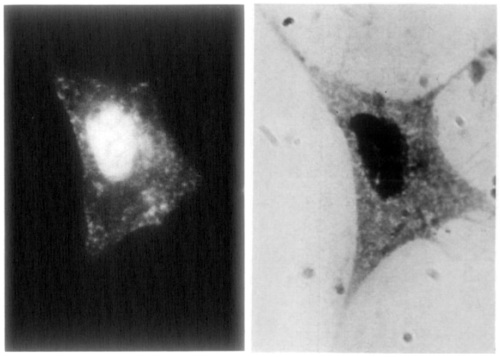
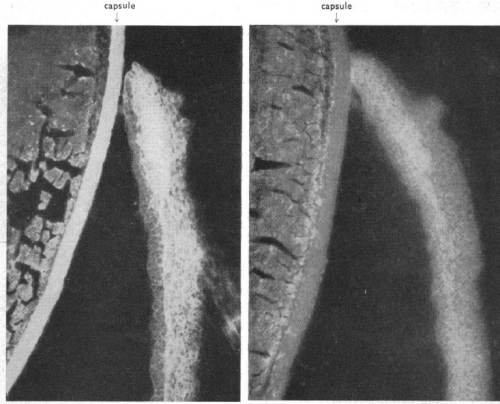
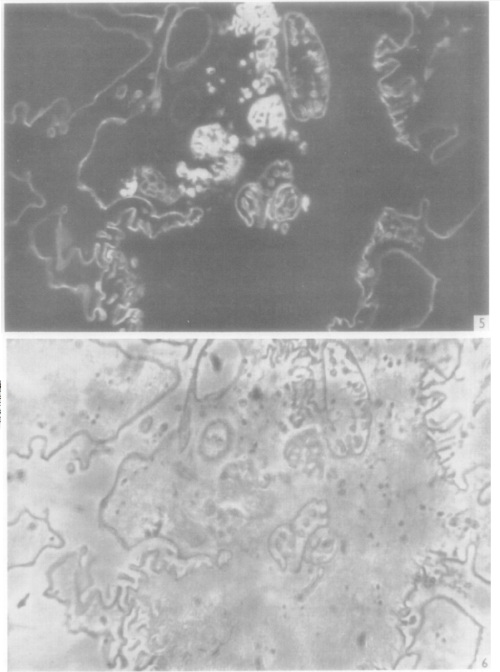
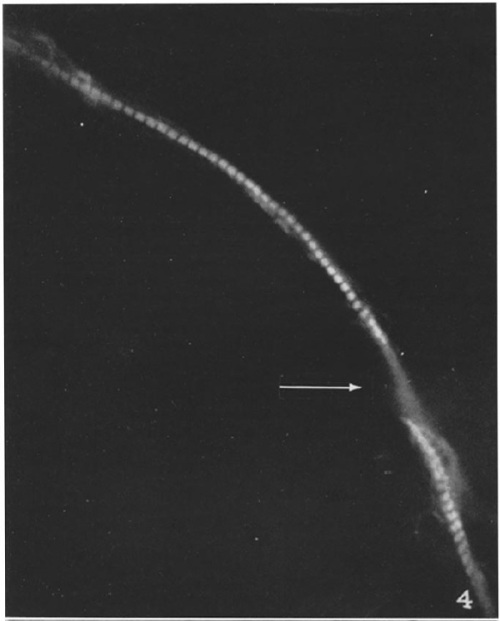
You can also use labeled antibodies to see things under the electron microscope (immuno-electron microscopy or immuno-EM). In 1960 Rifkin et al. (1) published an image of virus particles on a cell surface, labeled with ferritin-conjugated antibodies. As you can see, instead of the labeled antibodies changing the color of a region of the cell, each individual labeled antibody is visible as a “granule”.
This was made possible by an innovation published one year earlier, entitled Preparation of an electron-dense antibody conjugate (2). Ferritin is a small protein which just about all organisms use as an iron carrier. When “iron-loaded”, almost a quarter of its mass is iron atoms. Therefore this is an especially electron-dense molecule, visible as a dark spot under the electron microscope, as seen above. Soon, further advances let scientists see ferritin-labeled structures inside cells.
* * *
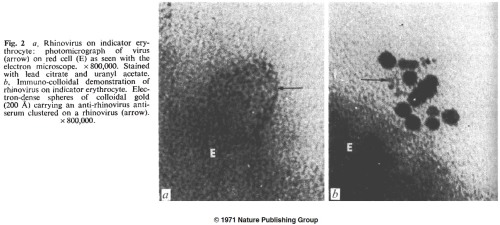
For about a decade ferritin was the label of choice for immuno-electron microscopy. Then in 1971, a new technique came along (3, 4), in which antibodies were mixed with a solution of colloidal gold until they absorbed to the metal’s surface. Gold-labeled antibodies could be separated from free antibodies by centrifugation. Immunogold is still the dominant immuno-EM staining method 40 years later.
* * *
In the 1960s, other techniques were created for immuno-EM. I have almost no EM experience and don’t know the pros and cons, but clearly there was a desire to get rid of the protein element of the electron-dense antibody label, and just attach the antibody to a metal ion. The protein was unnecessarily big, and subject to denaturation. So the 1960s also saw a lot of papers using antibodies labeled with mercury (technically the diazonium salt of tetraacetoxymercuriarsanilic acid (5) and p-(aminophenyl)-mercuric acetate (6), phrases which mean little to me).
There were also studies using antibodies labeled with heavier metals. In his long career, Ludwig Sternberger and his lab (at Johns Hopkins and elsewhere) invented several microscopy techniques, with the most important probably being the horseradish peroxidase (see original paper (7), and appreciation of it as a “citation classic”).
He also spent much of the 1960s devising improved metal-based antibody labels for electron microscopy, including immunouranium (8), immunouranium with added osmium for enhanced contrast (9), and finally immuno-diazothioether-osmium tetroxide (10), or immuno-DTO. The uranium methods seem somewhat useful, but as far as I can tell were only used by Sternberger’s own lab. Immuno-DTO in particular seemed almost unusable; they used it more than once, but a Pubmed search for “Immuno-DTO” only returns one result (11). As Sternberger himself says in a review (12) of his and other techniques:
Unfortunately, the diazotized diazothioethers are not very stable even at Dry Ice temperatures and the solid deteriorates in a few days. Therefore, it was not surprising to observe that the osmnium tetroxide-binding iower of diazothioether antibodies was unstable, even on storage in liquid nitrogen.
Oh well, it was a nice idea.
* * *
1. Rifkind RA, Hsu KC, Morgan C, Seegal BC, Knox AW, Rose HM (1960). Use of Ferritin-Conjugated Antibody to Localize Antigen by Electron Microscopy. Nature 187:1094-1095.
2. Singer SJ (1959). Preparation of an Electron-dense Antibody Conjugate. Nature 183:1523-1524.
3.Faulk WP, Taylor GM (1971). An Immunocolloid Method for the Electron Microscopy. Immunochemistry 8(11):1081-1083.
4. Faulk WP, Vyas GN, Phillips CA, Fudenberg HH, Chism K (1971). Passive Haemagglutination Test for Anti-rhinovirus Antibodies. Nature New Biology 231:101-104.
5. Pepe FA (1961). The Use of Specific Antibody in Electron Microscopy: I: Preparation of Mercury-Labeled Antibody. J Biophys Biochem Cytol 11(3):515-520.
6. Zhdanov VM, Azadova NB, Kulberg AY (1965). The Use of Antibody Labeled with an Organic Mercury Compound in Electron Microscopy. J Histochem Cytochem 13(8):694-687.
7. Sternberger LA, Hardy PH Jr, Cuculis JJ, Meyer HG (1970). The Unlabeled Antibody Enzyme Method of Immunohistochemistry: Preparation and Properties of Soluble Antigen-Antibody Complex (Horseradish Peroxidase-Antihorseradish Peroxidase) and its Use in Identification of Spirochetes. J Histochem Cytochem 18(5):315-333.
8. Donati EJ, Figge FHJ, Sternberger LA (1965). Staining of Vaccinia Antigen by Immunouranium Technique. Exp Mol Pathol 4(1):126-129.
9. Sternberger LA, Hanker JS, Donati EJ, Petrali JP, Seligman AM (1966). Method for Enhancement of Electron Microscopic Visualization of Embedded Antigen by Bridging Osmium to Uranium Antibody with Thiocarbohydrazide. J Histochem Cytochem 14(10):711-718.
10. Donati EJ, Petrali JP, Sternberger LA (1966). Formation of Vaccinia Antigen Studied by Immunouranium and Immuno-diaxothioether-osmium tetroxide Techniques. Exp Mol Pathol Apr:Suppl 3:59-74.
11. Sternberger LA, Donati EJ, Hanker JS, Seligman AM (1966). Immuno-diazothioether-osmium tetroxide (immuno-DTO) technique for staining embedded antigen in electron microscopy. Exp Mol Pathol Apr:Suppl 3:36-43.
12. Sternberger LA (1967). Electron Microscopic Immunocytochemistry: A Review. J Histochem Cytochem 15(3):139-159.